Researchers at the University of Delaware have discovered that an ancient gene — ribonucleotide reductase (RNR), which occurs in all cellular life — provides important biological insights into the characteristics of unknown viruses in the sea.
The results of this finding could ultimately lead to new tools for understanding the inner workings of marine microbial communities.
Eric Wommack, principal investigator on the project and a professor who holds joint appointments in the departments of Plant and Soil Sciences and Biological Sciences and the College of Earth, Ocean, and Environment, explained, “The reason we care about these inner workings is because of the influence of natural, microbial communities on the balance of carbon and nutrients in ecosystems. In order for us to understand how we’re impacting the natural systems of the planet, it all comes down to the movement of carbon and nutrients through systems. Anywhere there are viruses infecting bacteria, which is everywhere, this is a possible tool that can be used.”
Much of the work was conducted by Eric Sakowski, a doctoral student in the Department of Biological Sciences, along with a team of graduate and undergraduate students.
The results of the study will be published in an upcoming issue of the Proceedings of the National Academy of Sciences (PNAS).
“This research shines a light on viral ‘dark matter’ — a major challenge to understanding the diversity of life on Earth,” says Matt Kane, program director in the National Science Foundation’s (NSF) Division of Environmental Biology, which co-funded the research with NSF’s Division of Ocean Sciences. “The identification of ribonuclotide reductase as a marker for studying viral diversity and ecology is an important new tool for revealing how viruses affect microbial interactions and processes.”
Biological classifications
Wommack said that one of the key things biologists do in order to understand how organisms function is to develop classification schemes — categorizing everything and then figuring out what those categories mean.
In the modern era, biologists do not have to rely simply on what they can see or measure, as new technologies allow them to examine genes.
“Looking at a sequence of DNA, it’s kind of like a record of the evolutionary changes that have happened among the organisms or viruses that have that gene,” said Wommack.
Using sequence data, researchers are able to do a lot more connecting of the dots, such as understanding all the little changes in DNA that happened between humans and the last non-human ancestor.
RNR
Wommack said this study, which began in 2011, is “distilling viral biology down to the level of this single RNR gene, which is a very ancient gene. We’re talking, possibly before even the first cell. This gene is believed to have been critical to the emergence of DNA-based life.”
Sakowski added that the gene is “so ancient that every cell on the planet that we know of has it.”
A lot of viruses also have the gene but the problem with studying environmental viruses is that most of them are completely unknown.
“When we’re studying these viruses, they aren’t viruses that we can observe. We can’t grow them in the lab, we can’t physically look at most of them, so the only thing we have to go on is the genetic sequence. And then, if you don’t have sequence data that you’ve seen before, it’s really hard to make conclusions about the virus that it came from,” said Sakowski.
That’s where the researchers turned to RNR.
Viral predictions
Because RNR is so old and critical to DNA replication — which means that it will be important to viruses — Wommack and Sakowski felt that the RNR gene would be a good choice for trying to connect its evolutionary history with viral biology.
“We are always looking for ways to use sequence data to describe something about the biology of the viruses that we see and answer some baseline questions, such as which host do they infect? How fast do they grow? Does the sequence connect with their physical shape?” said Wommack.
Sakowski looked at RNR sequences from known viruses to see if there were any biological trends that would provide context to the unknown viruses. Using this marker gene approach, he found several things using RNR sequences.
“A marker gene, like RNR, is a specific gene that we use to understand evolutionary relationships and the community structure of the viral populations that we find in the environment,” said Sakowski. “The first nice thing about RNR is that we were able to take a look at a much broader spectrum of the viruses that are present and see which ones are the most abundant, not just within one small group but across a breadth of diversity. This includes the most abundant, and we think, the most biologically important viruses in the environment. So we can get a better understanding of what these natural populations look like.”
With RNR in marine environments they found it mostly occurs in lytic viruses — viruses that replicate rapidly — that the researchers believe are the key players in controlling a lot of the bacterial populations and influencing things like energy flow and carbon and other nutrient cycles.
They have also found that by comparing the RNR gene sequences of known viruses with those from environmental viruses, they can make predictions about the physical shape of some environmental viruses. They can also predict more about the biology of those viruses.
“RNR is really interesting because there are three different classes based on how the enzyme reacts to oxygen,” said Sakowski. “There’s an oxygen-dependent enzyme that only works when there’s oxygen available, an oxygen-independent version that can work regardless of whether oxygen is present or absent, and an oxygen-sensitive version of the enzyme that only works in the absence of oxygen.”
By knowing which version of the RNR gene the virus is carrying, they know the conditions under which it is actively replicating so they can tell if it is infecting a host that is potentially an aerobe — a host that’s using oxygen — or if it is replicating in a host that is an anaerobe.
“We can really start to gain insight into some of those biological aspects that you couldn’t with some of the other genes that we’ve been looking at,” said Sakowski.
Womamck said that predicting which host groups these viruses may be infecting has been a “difficult question to answer, and an incredibly important one because we’re only looking at half the equation when we look at the virus. Obviously some host population was infected and killed to produce those viruses so we believe from the analysis that for some groups we can actually predict the host groups that produce that virus.”
The implications for the field of microbial ecology and environmental microbiology are that researchers can now go in and look at a viral community and potentially, over time, make predictions about which host populations could be most impacted by viral infection.
“What it means is that we’re dissecting the inner workings of the system, which is ultimately what science is aiming to do, to really understand how natural systems work,” said Wommack.
Now that the evolutionary analysis has been done and the drawing of connections with biological features has been completed, Wommack said they hope more researchers will use this information going forward.
The researchers were aided along the way by undergraduate researchers, Mara Hyatt and William Kress, who worked with Sakowski and Wommack through the support of undergraduate research internships from Experimental Program to Stimulate Competitive Research (EPSCoR) program and the Delaware Water Resources Center (DWRC). Sakowski was funded on a graduate fellowship through the DWRC
They also acknowledged the Delaware IDeA Networks of Biomedical Research Excellence (INBRE) program and the Gordon and Betty Moore Foundation (through grant GBMF2732), both of which were instrumental in the research.
Article by Adam Thomas
Photos and images by Lindsay Yeager

Eric Sakowski at work in the laboratory.
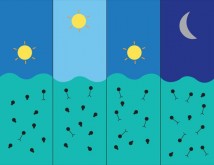
The infection dynamics of viruses infecting photosynthetic bacteria could be predicted from RNR sequences. In the morning there was greater production of short-tailed podoviruses, whereas, in the evening there was greater production of long-tailed myoviruses.